Bioinformatics Analysis on Ribulose-1,5-bisphosphate Carboxylase/ Oxygenase Large Subunits in Different Plants 
2. Rice Research Institute, Jiangxi Academy of Agricultural Sciences, Nanchang, 330200, P.R. China
1. Institute of Quality Safety and Standards of Agricultural Products Research, Jiangxi Academy of Agricultural Sciences, Nanchang, 330200, P.R. China
2. Rice Research Institute, Jiangxi Academy of Agricultural Sciences, Nanchang, 330200, P.R. China
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2011, Vol. 2, No. 15 doi: 10.5376/mpb.2011.02.0015
Received: 29 Sep., 2011 Accepted: 20 Oct., 2011 Published: 26 Oct., 2011
Zhang et al., 2011, Bioinformatics Analysis on Ribulose-1,5-bisphosphate Carboxylase/ Oxygenase Large Subunits in Different Plants, Molecular Plant Breeding Vol.2 No.15 (doi: 10.5376/mpb.2011.02.0015)
The nucleotide sequences and amino acid sequences of ribulose-1,5-bisphosphate carboxylase/oxygenase large subunits (rbcL) from Zea mays, Arabidopsis thaliana, Pisum sativum, Citrus sinensis, Phalaenopsis aphrodite subsp. formosana, emphasizing on Oryza sativa subsp. japonica, which were registered in the Genebank, were analyzed by the tools of bioinformatics in the following aspects: the composition and the physical and chemical characteristics of nucleotide sequences and amino acid sequences, signal peptide, trans-membrane topological structure, hydrophobicity or hydrophilicity, secondary structure, sequences comparisons, and molecular systemic evolution. The results were showed as follows: the amino acid composition and characteristics of rbcLs from different higher plants were approximately identical; signal peptide and trans-membrane topological structure were not detected in the rbcLs that exhibit the traits of hydrophilic protein; the secondary structure of rbcL manly constituted with α-helix and random coil; the nucleotide sequences and amino acid sequences possess high homologies; the rbcL DNA sequence can reflect the sibships among various higher plants clearly.
Ribulose-1,5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39, RuBisCo), transforming the carbon dioxide and ribulose-1,5-bisphosphate (RuBP) into two molecular 3-phosphoglyceric acid, catalyzes the first reaction of carbon dioxide fixation in photosynthetic dark reaction. Also, RuBisCo catalyzes the reaction of oxygen and RuBP to phosphoglyceric acid and phosphoglycollic acid, which is the first reaction of photorespiration. Therefore, RuBisCo is the key enzyme deciding the photosynthetic efficiency by regulating photosynthesis and photorespiration. Base on the dissimilarity of the primary and quarternary structures, the RuBisCos can be partitioned into three types: I form exists in higher plants and most prokaryotes, consisting of eight large subunits (50~60 kD) and eight small subunits (12~18 kD), presenting square symmetry structure (L8S8) (Andersson et al., 1989); â…¡ form was discovered in purple non-sulfur photosynthetic bacteria, and composed of only two large subunits (L2); â…¢ form was dug out in Thermococcus kodakaracinsis lately by Kitano (Kitano et al., 2001), likewise formed with only large subunits, and no small subunit, appearing structure of (L2)5. The RuBisCo large subunit (rbcL) gene of higher plants sets in the chloroplast DNA, and it is translated by chloroplast ribosome. On the contrary, the RuBisCo small subunit (rbcS) is synthesized in cytoplasm 80S ribosome because the gene exists in cell nucleus genome, and then transfers to chloroplast as precursor protein to assemble with large subunit after processed (Ellis, 1987; Roy, 1989). To date, the rbcL gene has been cloned from a great many plants, such as Oryza sativa subsp. Japonica (Hiratsuka et al., 1989), Zea mays (Maier et al., 1995), Nicotiana tabacum (Shinozaki et al., 1986), Arabidopsis thaliana (Sato et al., 1999), Citrus sinensis (Bausher et al., 2006), Phalaenopsis aphrodite susp. Formosana (Chang et al., 2006), Astragalus mongholicus (Guo et al., 2010), Marchantia polymorpha (Ohyama et al., 1986; 1988), Picea abies (Relle et al., 1995).
In this study, the rbcL nucleotide sequences and deduced amino acid (AA) sequences from various higher plants were analyzed by the tools of bioinformatics, expecting to provide some theoretical reference for further studies on plant RuBisCo. The plants include Zea mays, Arabidopsis thaliana, Pisum sativum, Citrus sinensis, Phalaenopsis aphrodite susp. formosana, and so on, emphasizing Oryza sativa subsp. japonica. And the following aspects were involved in the analysis: the compositions and the physical and chemical characteristics, signal peptide, transmembrane topological structure, hydrophobicity or hydrophilicity, secondary structure, sequence comparison, and molecular systemic evolution.
1 Results
1.1 The compositions and the physical and chemical characteristics analysis of rbcL nucleotide sequences and deduced amino acid sequences from plants
The compositions and the physical and chemical characteristics of rbcL nucleotide sequences and deduced AA sequences were discribed by ORF Finder, DNAstar, ProtParam and pI/Mw. The analyzed rbcLs data were derived from Oryza sativa subsp. japonica, Zea mays, Arabidopsis thaliana, Pisum sativum, Citrus sinensis, Phalaenopsis aphrodite subsp. formosana. All the initiation codons of the rbcLs genes are ATG, and the termination codons are TAG or TAA. The lengths of ORFs are about 143 4 bps, and the encoding proteins are approximately 477 AA residues. The molecular weight and theoretical isoelectric point of the polypeptides are similar among different plants. The proportions of acidic AA, alkaline AA, total electric AA, polar AA and hydrophobic AA in the total AA residues of the rbcLs show tiny differences. On the whole, the most abundant AA residues are Gly, Ala, Leu, Glu and Val. The rbcLs of Pisum sativum and Citrus sinensis belong to stable protein, while that of the other three plants are unstable protein, but the instability indexes of all rbcLs are close to 40% (Table 1).
|
Table 1 Comparison of compositions and physical and chemical characteristics of nucleotide sequences and deduced amino acid sequences of RuBisCo large subunits among different higher plants
|
1.2 The signal peptide analysis of plant rbcLs
The rbcL AA sequence signal peptide of Oryza sativa subsp. japonica was predicted by SignalP Server v. 3.0 online program (Nielsen et al., 1997; Bendtsen et al., 2004). The analysis was performed using Neural Networks Model (NN) method. The top values of original shearing site (C score), signal peptide (S score), and synthesized shearing site (Y score) are 0.059, 0.133, and 0.021, which locate at the 24th, 4th, and 8th AA residues, respectively (Figure 1). All the scores are far less than the critical threshold. Moreover, the probability of presence of signal peptide in the analysis of polypeptide applying Hidden Markov Models (HMM) method is zero. Therefore, it indicates that no signal peptide shearing site exists in the rbcL polypeptide. The similar results were observed in the prediction of rbcL AA sequences of Zea mays, Triticum aestivum, Arabidopsis thaliana, Citrus sinensis. Accordingly, it was inferred that the rbcLs polypeptide synthesized in higher plants chloroplast don't require to be protein transmembrane transfered.
1.3 The transmembrane topological structure analysis of plant rbcLs
The rbcL AA sequence transmembrane topological structure of Oryza sativa subsp. japonica was explored applying TMHMM Server v. 2.0 program (Ikeda et al., 2002). The total rbcL polypeptide locates outside the membrane (Figure 2), namely, it is absent of transmembrane topological structure in the rbcL AA sequence of Oryza sativa subsp. japonica. The same results can be gained from the analysis of rbcL AA sequences of Zea mays, Lolium perenne, Oncidium Gower Ramsey, Calycanthus floridus var. glaucus, Phalaenopsis aphrodite subsp. formosana, and so on.
|
Figure 2 Transmembrane topological structure analysis of rice rbcL
|
1.4 The hydrophobicity and hydrophilicity analysis of plant rbcLs
The hydrophobicity and hydrophilicity analysis of the rbcL AA sequence of Oryza sativa subsp. japonica was fulfilled with ProtScale program (Kyce and Doolittle, 1982). The most hydrophilic AA residue in the polypeptide is Asn, located at 306th, because of the lowest score of -2.644. And the most hydrophobic AA residue is Ala, situated at 378th, which has the top score of 1.778. As for the whole polypeptide, the hydrophobic and hydrophilic AA residues distribute uniformly, but the number of hydrophilic AA residues is higher than that of hydrophobic AA residues, and any obvious hydrophobic AA residues concentrative region can't be detected (Figure 3). Similar distributive rule of hydrophobic and hydrophilic AA residues was found in other rbcL AA sequences from Nicotiana tabacum, Lolium perenne, Medicago truncatula, Pisum sativum, and Citrus sinensis. Thus, the results implies that the rbcLs in higher plants are hydrophilic protein, which is in accord with the previous conclusion that transmembrane topological structure is absent in rbcLs of higher plants.
1.5 The rbcL secondary structure analysis of plants
The rbcL polypeptide secondary structure of Oryza sativa subsp. japonica was detected with SOPMA (Geourjon and Deléage, 1995). Alpha-helix and random coil are the principal structural elements in rbcL polypeptide of Oryza sativa subsp. japonica, and extended strand and β-turn occupy a little scale, which intersperse among the whole protein (Figure 4). According to the statistic assay consequence, the proportions of α-helix, extended strand, β-turn and random coil in the rbcL secondary structural components of Oryza sativa subsp. japonica are 40.25%, 16.56%, 9.85% and 33.33%, respectively. Likewise, similary distributive regularity of the four kinds of secondary structural units in rbcL has been detected in many other higher plants, such as Zea mays, Triticum aestivum, Nicotiana tabacum, Medicago truncatula, Oncidium Gower Ramsey, Pisum sativum, Citrus sinensis.
1.6 Comparisons among the rbcL nucleotide sequences and deduced amino acid sequences from different plants
The homological comparisons of rbcL nucleotide sequences and deduced AA sequences were accomplished by online tool BLAST in NCBI (Alcschul et al., 1997). The results indicate high homologies of rbcL nucleotide sequences between Oryza sativa subsp. japonica and other higher plants which are comprised of Triticum aestivum, Nicotiana tabacum, Lolium perenne, Medicago truncatula, Oncidium Gower Ramsey, Calycanthus floridus var. glaucus, Podocarpus macrophyllus. The identities are 96%, 88%, 96%, 87%, 91%, 88%, 85%, respectively. The identities of further deduced AA sequence comparisons using BLASTp are 97%, 93%, 97%, 93%, 96%, 94%, 94%, respectively, while the similarities of that are separately 99%, 97%, 99%, 97%, 98%, 97% and 98%. Relative to the results of nucleotide sequence comparisons by BLASTn, higher similarities can be discovered in the AA sequence comparisons between Oryza sativa subsp. japonica and other six plants. Also, analogous results were obtained in the comparisons of rbcL nucleotide sequences and deduced AA sequences between Oryza sativa subsp. japonica and a great number of other higher plants, such as Zea mays, Arabidopsis thaliana, Pisum sativum, Citrus sinensis, Phalaenopsis aphrodite subsp. formosana. The regularity, which exhibited higher and broader similarities in rbcL AA sequences than in nucleotide sequences among various plants, is supported by above-mentioned results.
The multiple alignment analysis of rbcL AA sequences among Cathaya argyrophylla, Calycanthus floridus var. glaucus, Nicotiana tabacum, Solanum lycopersicum, Citrus sinensis, Arabidopsis thaliana, Allium cepa, Phalaenopsis aphrodite subsp. formosana, Oryza sativa subsp. japonica and Zea mays was carried out using Clustalx (Higgins and Sharp, 1988; 1989; Thompson et al., 1997; Jeanmougin et al., 1998) and DNAMAN software. Super identities of rbcL AA sequences were illustrated among higher plants, regardless of between gymnosperm and angiosperm, or between dicotyledon and monocotyledon (Figure 5). It was demonstrated that there are exceeding conservatism and homologies among higher plant rbcLs.
1.7 The molecular systemic evolution analysis on rbcL DNA sequences from plants
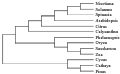
The homologous evolutionary relationship among rbcL DNA sequences from Nicotiana tabacum, Solanum lycopersicum, Spinacia oleracea, Arabidopsis thaliana, Citrus sinensis, Calycanthus floridus var. glaucus, Phalaenopsis aphrodite subsp. formosana, Oryza sativa subsp. japonica, Saccharum officinarum, Zea mays, Cycas taitungensis, Cathaya argyrophylla, and Pinus thunbergii, was performed by Clustalx and MEGA5 software (Saitou and Nei, 1987; Tamura et al., 2004), using Neighbor-Joining (NJ) method. The rbcL DNA sequences, derived from thirteen higher plants, were assembled into two big clusters. One group is comprised of Cycas taitungensis, Cathaya argyrophylla, Pinus thunbergii, and other ten plants constitute the other group. All the thirteen plants originate from a common ancestor. In the phylogenetic tree, the connection of the branches reflects the rbcLs evolutionary relationship of different plants clearly. The three gymnospermous plants, containing Cycas taitungensis, Cathaya argyrophylla and Pinus thunbergii, constitute a branch, which is distinguished from the other branch that include other ten plants, belonging to angiosperm. And the ten angiospermous plants can be further divided into two branches of dicotyledon and monocotyledon. Also, the Solanaceae plant, containing Nicotiana tabacum and Solanum lycopersicum, and the Poaceae plant, including Oryza sativa subsp. japonica, Zea mays, and Saccharum officinarum, constitute two small branches, respectively, on account of their close relationship (Figure 6).
The molecular level evolutionary relationship was applied in biological systemic taxology widespreadly, after the advance of "molecular evolutionary clock" and "neutral theory" in 1960s. A few divergences are present in the application of molecular evolution to biological taxonomy, due to the dispute of "constant speed of sequence evolution" and "darwinian positive selection" in academic world. However, it is acknowledged that the evolutionary units above family can be differentiated exactly with the phylogenetic analysis of DNA and AA sequence, which was proved adequately in this study. The Zea mays and Saccharum officinarum are separated from Oryza sativa subsp. japonica correctly (Figure 6), in virtue of their closer relationship, even though all the three plants belong to Gramineae.
2 Discussion
In this study, we demonstrated that the rbcLs from different higher plants don't possess signal peptide, transmembrane topological structure and the traits of hydrophobic protein. The principal secondary structural elements are α-helix and random coil. The compositions and the physical and chemical characteristics are similar, and extremely high homologies were exhibited among different higher plants. The evolutionary relationship reflected by DNA sequences corresponds with traditional botanical taxonomy.
It is known that the sequences and structures of rbcLs from different higher plants get high homologies, and the similarities of that are above 80%, while the similarities of rbcSs are much smaller and less than 50%. All the analyzed rbcL ORFs from higher plants are about 1 434 bp, and translate into polypeptides that consist of nearly 477 AA residues (Table 1). The similarities of the rbcL AA residues from different higher plants are more than 97%, and the inferior homological region in the rbcL polypeptide mainly locates at the C-terminal (Figure 5). The high homology of rbcLs indicates the importance of structural stability in maintaining high catalytic efficiency. Also, it implies that the overwhelming majority of rbcL AA residues play a crucial role in keeping the structural stability, as the report that the RuBisCo catalytic efficiency can be altered obviously when some AA residues of rbcL were substituted (Chen et al., 1988; 1993; Seokjoo and Robert., 1997; Bainbridge et al., 1998; Pippa et al., 1998).
As a double functional enzyme, RuBisCo catalyzes the oxygenation reaction of RuBP when it is catalyzing the carboxylation reaction of that. Because of the characteristics of RuBisCo, the plant will suffer a great loss of about 20%~50% of the organic carbon, fixed by the carboxylation reaction, no merely energy (Li et al., 2001). So in theoretically, the improvement of crop RuBisCo is a breakthrough point in crop variety improvement using modern biotechnology, and has a tempting perspective (Mann, 1999; Parry et al., 2007). Up to now, rapid progress has been making in studies on RuBisCo structures, biological functions and regulations, and enzymatic characters, but it is still theoretical in improving crop photosynthetic efficiency and increasing yield via the modification of RuBisCo. Therefore, further exploration of RuBisCo natures and molecular characteristics are indispensable to lay a solid foundation of enhancing crop RuBisCo catalytic efficiency and increasing the photosynthetic output, for instance, the diversity of RuBisCo structures and functions among different plants, environmental regulations and active mechanisms, and the relationship of protein structures and functions.
Authors' contributions
SMZ and MN have finished the paper, GKA and RR also read the manuscript and revised it. All authors had read and consented the final text.
Acknowledgements
We thank Dr. Nazir Ahmad Bhat, Sr. Scientist, SKUAST-K Srinagar for his help in preparation of manuscript.
References
Alcschul S.F., Madden T.L., Schffer A.A., Zhang J.H., Zhang Z., Miller W., and Lipman D.J., 1997, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res., 25(17): 3389-3402http://dx.doi.org/10.1093/nar/25.17.3389 PMid:9254694 PMCid:146917
Andersson I., Knight S., Schneider G., Schneider G.,Lindqvist Y., Lundqvist T., Brändén C.I., and Lorimer G.H., 1989, Crystal structure of the active site of ribulose-1,5- bisphosphate carboxylase, Nature, 337(19) : 229-234 http://dx.doi.org/10.1038/337229a0
Bainbridge G., Anralojc P.J., Madgwick P.J., Pitts J.E., and Parry M.A., 1998, Effect of mutation of lysine-128 of the large subunit of ribulose bisphosphate carboxylase/oxygenase from Anacystis nidulans, Biochem J, 336(Pt2): 387-393 PMid:9820816 PMCid:1219883
Bausher M.G., Singh N.D., Lee S.B., Jansen R.K., and Daniell H., 2006, The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var 'Ridge Pineapple: organization and phylogenetic relationships to other angiosperms, BMC Plant Biol., 6: 21-30 http://dx.doi.org/10.1186/1471-2229-6-21 PMid:17010212 PMCid:1599732
Bendtsen J.D., Nielsen H., von Heijne G., and Brunak S., 2004, Improved prediction of signal peptides: SignalP 3.0, J. Mol. Biol., 340(4): 783-795 http://dx.doi.org/10.1016/j.jmb.2004.05.028 PMid:15223320
Chang C.C., Lin H.C., Lin I.P., Chow T.Y., Chen H.H., Chen W.H., Cheng C.H., Lin C.Y., Liu S.M., ChangC.C., and Chaw S.M., 2006, The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications, Mol. Biol. Evol., 23(2): 279-291 http://dx.doi.org/10.1093/molbev/msj029 PMid:16207935
Chen Z., Chastain J.C., Alabed S.R., Chollet R., and Spreitzer R.J., 1988, Reduced CO2/O2 specificity of ribulose-bisphosphate carboxylase/oxygenase in a temperature-sensitive chloroplast mutant of Chlamydomonas, PNAS, 85: 4696-4699 http://dx.doi.org/10.1073/pnas.85.13.4696
Chen Z., Seokjoo H., and Robert J.S., 1993, Thermal instability of ribulose-l,5-bisphosphate carboxylase/oxygenase from a temperature-conditional chloroplast mutant of Chlamydomonas reinhardtii, Plant Physiol., 101: 1189-1194 PMid:12231772 PMCid:160638
Ellis R.J., 1987, Protein as molecular chaperons, Nature, 328: 378-379 http://dx.doi.org/10.1038/328378a0 PMid:3112578
Geourjon C., and Deléage G., 1995, SOPMA: Significant improvement in protein secondary structure prediction by consensus prediction from multiple alignments, Comput. Appl. Biosci., 11(6): 681-684 PMid:8808585
Guo H., Wang W., Yang N., Guo B.L., Zhang S., Yang R.J., Yuan Y., Yu J.L., Hu S.N., and Sun Q.S., 2010, DNA barcoding provides distinction between Radix astragali and its adulterants, Science China Life Science, 53(8): 992-999 http://dx.doi.org/10.1007/s11427-010-4044-y PMid:20821298
Higgins D.G., Sharp P.M., 1988, CLUSTAL: a package for performing multiple sequence alignment on a microcomputer, Gene, 73: 237-244 http://dx.doi.org/10.1016/0378-1119(88)90330-7
Higgins D.G., and Sharp P.M., 1989, Fast and sensitive multiple sequence alignments on a microcomputer, CABIOS, 5: 151-153 PMid:2720464
Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C.R., and Meng B.Y., 1989, The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals, Mol. Gen. Genet., 217(2-3): 185-194 http://dx.doi.org/10.1007/BF02464880
Ikeda M., Arai M., and Lao D.M., 2002, Transmembrane topology prediction methods: A reassessment and improvement by a consensus method using a dataset of experimentally characterized transmembrane topologics, In Silico Biol., 2(1): 19-33 PMid:11808871
Jeanmougin F., Thompson J.D, and Gouy M., 1998, Multiple sequence alignment with ClustalX, Trends Biochem. Sci., 23: 403-405 http://dx.doi.org/10.1016/S0968-0004(98)01285-7
Kitano K., Maeda N., Fukui T., Atomi K., Imanaka T., and Miki K., 2001, Crystal structure of a novel type archaeal rubisco with pentagonal symmetry, Structure, 9(6): 473-481 http://dx.doi.org/10.1016/S0969-2126(01)00608-6
Kyce J., and Doolittle R.F., 1982, A simple method for displaying the hydropathic character of a protein, J. Mol. Biol., 157(6): 105-132 PMid:7108955
Li Y.S., Li D.M., and Zhu Y.G., 2001, Progresses of molecular biology for super high yield in rice (in Chinese with English abstract), Research of Agricultural Modernization, 22(5): 283-288
Mann C.C., 1999, Genetic engineers aim to soup up crop photosynthesis, Science, 283(5400): 314~ 316 http://dx.doi.org/10.1126/science.283.5400.314 PMid:9925484
Maier R.M., Neckermann K., Igloi G.L., and Kössel H., 1995, Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing, J. Mol. Biol., 251(5): 614-628 http://dx.doi.org/10.1006/jmbi.1995.0460 PMid:7666415
Nielsen H., Engelbrecht J., Brunak S., and Heijne G., 1997, Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites, Protein Engineering, 10(1): 1-6 http://dx.doi.org/10.1093/protein/10.1.1
Ohyama K., Fukuzawa H., Kohchi T., Shirai H., Sano T., Sano S., UmesonoK., Shiki Y., Takeuchi M., Chang Z., Aota S.I., InokuchiH., and Ozeki H., 1986, Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA, Nature, 322: 572-574 http://dx.doi.org/10.1038/322572a0
Ohyama K., Fukuzawa H., Kohchi T., Sano T., Sano S., Shirai H., Umesono K., Shiki Y., Takeuchi M., Aota Z.C.S., Inokuchi H., and Ozeki H., 1988, Structure and organization of Marchantia polymorpha chloroplast genome. I. Cloning and gene identification, J. Mol. Biol. 203(2): 281-298 http://dx.doi.org/10.1016/0022-2836(88)90001-0
Parry M.A.J., Madgwick P.J., Carvahlo J.F.C., Andralojc P.J., 2007, Prospects for increasing photosynthesis by overcoming the limitations of Rubisco, J. Agric. Sci., 145(1): 31-43 http://dx.doi.org/10.1017/S0021859606006666
Pippa J.M., Saroj P., and Martin A.J.P., 1998, Effect of mutations of residue 340 in the large subunit polypeptide of Rubisco from Anacystis nidulans, Eur. J. Biochem., 253(3): 476-479 PMid:9654099
Relle M., Furst P., and Wild A., 1995, Short duplication in a cDNA clone of the rbcL gene from Picea abies, Plant Physiol., 107(3): 1037-1038http://dx.doi.org/10.1104/pp.107.3.1037 PMid:7716241 PMCid:157232
Roy H., 1989, Rubisco assembly: a model system for studying the mechanism of chaperonin action, The Plant Cell, 1(11): 1035-1042 http://dx.doi.org/10.2307/3869020 PMid:2577726 PMCid:159840 http://dx.doi.org/10.1105/tpc.1.11.1035 PMid:2577726 PMCid:159840
Saitou N., and Nei M., 1987, The neighbor-joining method: A new method for reconstructing phylogenetic trees, Molecular Biology and Evolution, 4(4): 406-425 PMid:3447015
Sato S., Nakamura Y., Kaneko T., Asamizu E., and Tabata S., 1999, Complete structure of the chloroplast genome of Arabidopsis thaliana, DNA Res., 6(5): 283-290 http://dx.doi.org/10.1093/dnares/6.5.283 PMid:10574454
Seokjoo H., and Robert J.S., 1997, Complementing substitutions at the bottom of the barrel influence catalysis and stability of sibulose-bisphosphate carboxylase/oxygenase, The Journal of Biological Chemistry, 272(17): 11114–11117 http://dx.doi.org/10.1074/jbc.272.17.11114 PMid:9111007
Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K., Ohto C., Torazawa K., Meng B.Y., Sugita M., Deno H., Kamogashira T., Yamada K., Kusuda J., Takaiwa F., Kato A., Tohdoh N., Shimada H., and Sugiura M., 1986, The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression, EMBO J., 5(9): 2043-2049 PMid:16453699 PMCid:1167080
Tamura K., Nei M., and Kumar S., 2004, Prospects for inferring very large phylogenies by using the neighbor- joining method, PNAS, 101(30): 11030-11035
http://dx.doi.org/10.1073/pnas.0404206101 PMid:15258291 PMCid:491989
Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., and Higgins D.G., 1997, The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools, Nucleic Acids Research, 25: 4876-4882 http://dx.doi.org/10.1093/nar/25.24.4876 PMid:9396791 PMCid:147148
. PDF(1167KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Biaojin Zhang
. Linguang Luo
. Xiangxi Zhang
. Ruili Li
. Yan Song
. Dawen Zhang
. Yuanyuan Nie
. Yanbing Zeng
. Qiegen Liao
. Yihua Wei
Related articles
. Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo)
. Plant
. Rice ( Oryza sativa L)
. Bioinformatics
Tools
. Email to a friend
. Post a comment
.png)



.png)